The art of Monte Carlo lies in one's ability to design as large a move as possible with change in the system's energy that is as small as possible. In the quest for the 'killer' moves, many of our designs that we hoped to result in a shortcut end up instead being a lengthy detour. I reported in this place ( Information Quarterly of CCP5 #36, 1993) one with the same title - hence the #2. I am presenting an other such detour here.
The standard move in molecular Monte Carlo consists of a combination of translation and rotation of a single molecule. Even this simple move allows a plethora of variants (small or large stepsizes, biasing in the direction of forces and torques, modulating the stepsize based in the energy or local density, etc.). However, to the best of my knowledge, there is no published report on limiting the move to either translation or rotation.
It occurred to me recently that it is possible that by always translating and rotating a molecule we might be doing ourselves a disservice and these two types of changes actually interfere with each other in some way, reducing the efficiency of the moves. To test this, I implemented into my Monte Carlo program MMC (available at https://mezeim01.u.hpc.mssm.edu/mmc) the option of alternating translations and rotations.
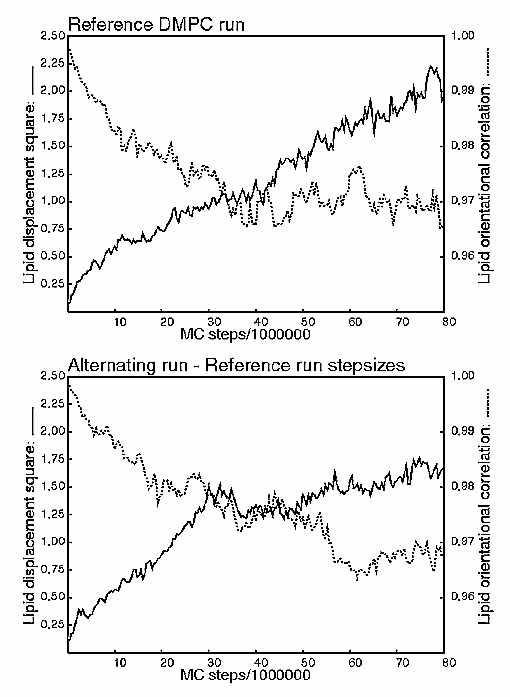
The new moves were tested on both liquid water and a solvated lipid bilayer (DMPC - see P. Jedlovszky & M. Mezei, J. Chem. Phys., 110, 2991-3002 (1999)). The sampling efficiency was monitored by the decay of the correlation between the initial and final orientation of the molecules and by the mean square displacement of the molecular centers. The results, however, bore out the wisdom of using combined translations and rotations. A number of stepsize combinations were tried for both methods, but the combined moves consistently outperformed the alternating moves although the differences were generally small. To illustrate the difference between the two runs the figures show the mean square displacements and decay of orientational correlation (orientation around the direction normal to the bilayer) as a function of the runlength for the lipid molecules.
So, it seems that the combined moves are here to stay. I did leave the option of alternating them in the program, though - it is useful for tuning the displacement and rotation stepsizes and there may exist some weird shaped molecule where alternating translations and rotations would outperform the combined move.
Incidentally, the figures also illustrate the difficulty of fine-tuned optimization of sampling parameters. Generally, the larger the limiting slope of the mean square displacement, the better the sampling. However, for nontrivial solutes like a lipid molecule this slope is hard to establish precisely enough for reliable comparison of two runs with similar characteristics.
